📋 Task
Based on the insights of the last two autoPET challenges, we expand the scope of the autoPET III challenge to the
primary task of achieving multitracer multicenter generalization of
automated lesion segmentation.
To this end, we provide participants
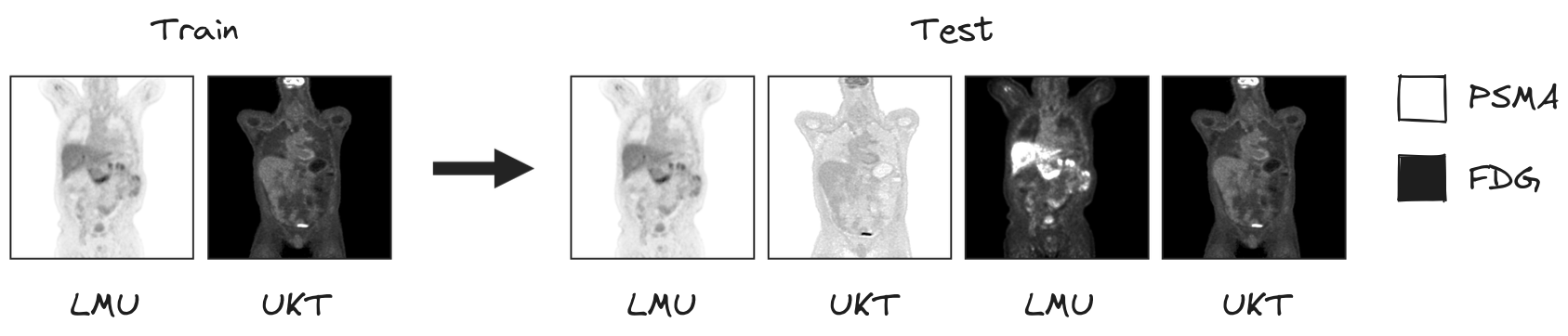
access to two different PET/CT training datasets: a large, annotated FDG-PET/CT dataset acquired at the University Hospital Tübingen, Germany (UKT), and a large, annotated PSMA-PET/CT dataset acquired at LMU Hospital, LMU Munich, Germany (LMU). The FDG-UKT dataset was already used in the autoPET I and II challenges. The PSMA-LMU dataset is new. It encompasses 597 PET/CT volumes of male patients diagnosed with prostate carcinoma from LMU Hospital and shows a significant domain shift from the UKT training data (different tracers, different PET/CT scanners, and acquisition protocols).
Algorithms will be tested on PSMA and FDG data from LMU and UKT, respectively. We will use a mixed model framework to rank valid submissions accounting for the effects of different tracers and different sites.
In addition, we will have a second award category, where participants are invited to submit our baseline model trained with their advanced data pipelines. This category is motivated by the observation that in autoPET I and II, data quality and handling in pre- and post-processing posed significant bottlenecks. Due to the rarity of publicly available PET data in the medical deep learning community, there is no standardized approach to preprocess these images (normalization, augmentations, etc.). The second award category will thus additionally promote data-centric approaches to automated PET/CT lesion segmentation.
Testing will be performed on 200 studies (held-out test database). Test data will be drawn in part (50%) from the same sources and distributions as the training data. The other part will be drawn crosswise from the other center, i.e. PSMA from Tuebingen (25%) and FDG from LMU (25%).
Goals of the challenge:
- Accurate segmentation of FDG- and PSMA-avid tumor lesions in whole-body PET/CT images. The specific challenge in automated segmentation of lesions in PET/CT is to avoid false-positive segmentation of anatomical structures that have physiologically high uptake while capturing all tumor lesions. This task is particularly challenging in a multitracer setting since the physiological uptake partly differs for different tracers: e.g. brain, kidney, heart for FDG and e.g. liver, kidney, spleen, submandibular for PSMA.
- Robust behavior of the to-be-developed algorithms with respect to moderate changes in the choice of tracer, acquisition protocol, or acquisition site. This will be reflected by the test data which will be drawn partly from the same distribution as the training data and partly from a different hospital with a similar, but slightly different acquisition setup.
📋 Award category 1: Best generalizing model
📋 Award category 2: Datacentrism
Best data-handling wins! In real-world applications, especially in the medical domain, data is messy. Improving models is not the only way to get better performance. You can also improve the dataset itself rather than treating it as fixed. This is the core idea of a popular research direction called Data-Centric AI (DCAI). Examples are outlier detection and removal (handling abnormal examples in dataset), error detection and correction (handling incorrect values/labels in dataset), data augmentation (adding examples to data to encode prior knowledge) and many more. If you are interested: a good resource to start is DCAI.
The rules are: Train a model which generalizes well on FDG and PSMA data but DO NOT alter the model architecture or get lost in configuration ablations. For that we will provide a second baseline container and a tutorial how to use and train the model. You are not allowed to use any additional data and the datacentric baseline model will be in competition. This means to be eligible for any award you need to reach a higher rank than the baseline. Models submitted in the datacentric category will also score in award category 1.
